Developments in the beverages markets are posing greater challenges to soft drink and beer manufacturers’ quality control. An unprecedented number of new products and product variants have become available to consumers. About 40% of soft drink products currently on sale in Europe were introduced during the past five years [1]. Many of the novel beverages impose lower antimicrobial hurdles on contaminants. Almost all marketed beverages have inherent properties (for example carbonation, alcohol and thermal pre-treatment) that slow or stop the growth of most potential contaminants. However, new soft drinks tend to possess higher levels of nutrients for microbial growth, lower acidity or milder carbonation levels than their traditional counterparts. Manufacturers have also cut down on thermal and chemical preservation. New functional and exotic additives as well as a more frequent usage of imported ingredients have expanded the range of potential spoilage organisms. For example, acetic acid bacteria of the genus Asaia have emerged as contaminants of flavoured mineral waters [2]. The increasing diversity of packaging solutions also poses additional challenges. For example, plastics vary considerably with respect their surface characteristics and gas permeability. In PET bottles, oxygen levels increase with time, whereas oxygen cannot permeate conventional glass bottles [3]. Recent improvements in reducing oxygen levels in beer processing and final packaging have improved the conditions for growth of anaerobic bacteria, which are now found more frequently in beer. All this calls for extra vigilance. Whenever beverage products are newly developed or modified, it is necessary to go through every change in the recipe, processing and packaging in order to consider the microbial – and, consequently, financial – risks. Soft drink lines may run at 30,000 bottles per hour, for perhaps 16 hours a day. Thus, in the event of a contamination, half a million items are lost per day until detection, with considerable reprocessing, disposal and – possibly – product recall costs to follow. Even in the usual case of no contamination being found that would require corrective action, significant product holding and storage costs are incurred until the microbiological results allowing the release of the product are obtained. Therefore, every day less of waiting until reliable results become available reduces both the risks and the costs. Traditional monitoring methods may require five days or more to generate results for spoilage yeasts, moulds or bacteria. Detection of contaminants that have adapted to a product may even take much longer. For example, Dekkera yeast strains that had been repeatedly subcultured in beer were found to exhibit diminished culturability on several recommended media, with detection times considerably extended [4]. Manufacturers have thus shown keen interest in reducing the time to result, particularly of slow-growing contaminants. To meet these new challenges, a fluorescent staining system for the quantitative detection of viable and culturable contaminants in filterable samples can be applied. The Milliflex® Quantum (EMD Millipore) platform identifies micro-colonies days before they become visible to the naked eye. Non-specific enzymes contained in all viable cells are able to cleave the added non-fluorescent viability marker, resulting in a fluorescent product that, in effect, vastly enhances the visibility of an emerging colony. Most importantly from a quality control perspective, the method is non-destructive: the detected micro-colonies can be cultivated to continue growing, yielding visible colonies that allow subsequent identification by means of any available technology. The basic procedure is to filter 100 mL of beverage sample through a pre-sterilised filtration funnel which is attached to a membrane cassette. When completed, the funnel is snapped off on a pre-filled agar plate, transferring the membrane cassette onto the plate. The bipartite entity is subsequently incubated. Because only micro-colonies need to emerge, the incubation period is much shorter than for visible colonies to grow. After incubation, the membrane cassette with its attached agar plate is taken from the incubator and its two constituents separated. The membrane cassette is then placed onto a staining cassette baring a sterile pad which needs to have been freshly saturated with a fluorescent staining solution. This entity is then placed into the incubator at 32.5°C. After 30 minutes it is transferred onto the reader instrument for counting of any stained colonies through the reader’s window or via a computer screen. If any micro-colonies are present, the membrane unit is removed, placed onto fresh agar media and re-incubated for later identification. To determine whether this system delivers results consistent with those of the traditional plate count method, soft drink and beer samples spiked with an appropriate dilution of typical contaminants were analysed according to both methods. The quantitative results and the time required to generate these results were compared.
Beverage samples
Soft drinks vary considerably with regards to their physical properties and ingredients. As a consequence a wide range of samples were selected for the assays: • Ice tea peach • Ice tea red • Ice tea green and grapefruit • Ready-to-drink black tea • Ready-to-drink green tea • Aromatised water • Energy drink • Apple juice • Strawberry juice • Mango juice All of these beverages proved adequately filterable, with the exception of the strawberry and mango juices, which were too viscous. To improve filterability, these were pre-treated by adding 150µL of a sterile pectinase solution (9500U/mL) to 1mL of the respective beverage and incubated for 30 minutes at 37°C. After incubation the samples were filtered with 10mL of sterile 1% Tween® 80 surfactant. To reduce the fluorescence background, they were subsequently rinsed twice, first with 100mL of sterile 1% Tween® 80 surfactant, then with 100mL of sterile physiological water. For the beer assays, three different products were selected, all of which proved filterable. To assess whether the beer exerted any antimicrobial effect that would necessitate preliminary rinsing, the samples, as well as a water control, were inoculated with a microbial suspension. As a percentage of the water control count (average of three replicates), the beer sample counts (average of three replicates each) reached 96.9% for Product A, 96.3% for Product B and 88.9% for Product C and thus considerably more than 70%, an acceptance threshold selected because the European and US Pharmacopoeias stipulate this level for pharmaceuticals. Thus these samples did not exert any antimicrobial activity that would have made any rinsing steps necessary.
Contaminant strains
For spiking, suspensions at around 20 to 80 CFU were prepared of the following yeast, mould and bacteria strains, all of which are known to cause spoilage in soft drinks and/or beer: Saccharomyces cerevisiae (ATCC® 7754). S. cerevisiae is a carbonation-tolerant, highly fermentative yeast. It is important for the brewing and baking industries but one of the most frequent spoilers of lemonades and fruit juices. Lactobacillus acidophilus (ATCC® 4356). L. acidophilus is found in the normal bacterial flora of humans but is also a spoilage organism of soft drinks. Zygosaccharomyces bailii (DSM 70492): Z. bailii is a highly fermentative yeast and notorious spoilage organism of soft drinks. It grows in high sugar concentrations and is extremely resistant to a variety of weak-acid preservatives. Wild strain of Candida parapsilosis. C. parapsilosis is not only a yeast that contaminates fruit juices but also a human pathogen, causing sepsis as well as wound and tissue infections in immuno-compromised patients. Spores of Aspergillus brasiliensis (ATCC® 16404). This A. brasiliensis mould strain, formerly known as A. niger, is a significant spoiler of fruit, causing the so-called ‘black mould’. Dekkera bruxellensis (wild type). D. bruxellensis is a carbonation and ethanol-tolerant, highly fermentative, slow-growing yeast and a contaminant of soft drinks and beer. Spoilage may not become apparent for weeks.
Growth conditions
To culture the contaminants, media recommended for beverage monitoring were used. For the soft drink assays these were SDA, MEA, PDA, Y&M, m-Green, Orange Serum, MRS and BAT, and for the beer assays Wallerstein agar. The strains were cultured at 30°C, except for L. acidophilus and D. bruxellensis, which were grown at 37°C and 27°C, respectively. To determine if any of the media used exhibited background fluorescence, the media cassettes were tested following the general procedure. No background fluorescence was detected.
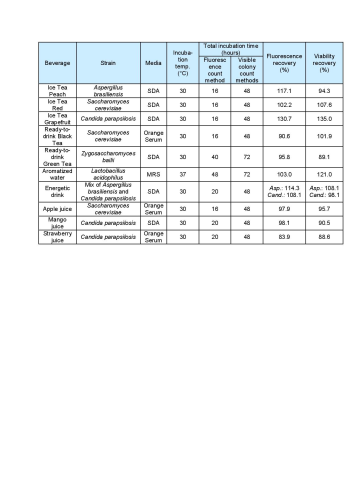
Results
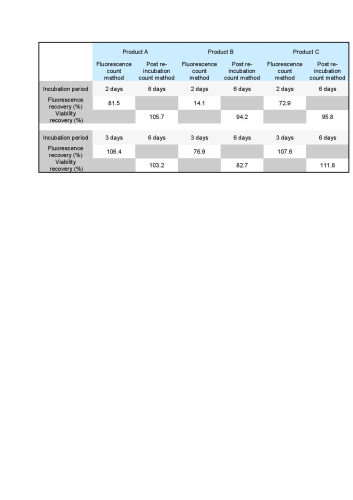
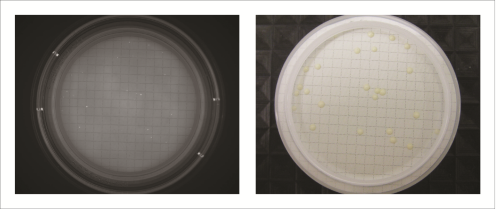
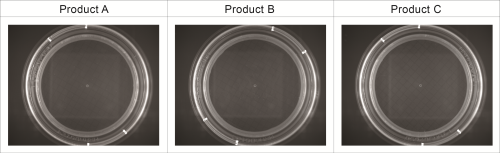
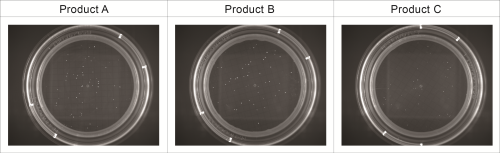
All recovery rates were determined on the basis of the average of three replicate plate counts. ‘Fluorescence recovery’ is the average fluorescent micro-colony count as a percentage of the average traditional method colony count. ‘Viability recovery’ is the average colony count after re-incubation as a percentage of the average traditional method colony count. For soft drinks, a variety of assays, crossing beverages, strains and media, were performed. For the traditional colony counts and the colony counts after re-incubation, plates were cultured for a total of between 48 and 72 hours, whereas for the fluorescence micro-colony count technique, the incubation period was between 16 and 48 hours. All assays resulted in fluorescence recovery rates greater than our acceptance threshold of 70% (see Table 1). This demonstrates that the fluorescence count method had sufficiently enabled detection of the micro-colonies that subsequently continued to grow into visible colonies. Viability recovery also exceeded 70% for all samples, with results ranging from 88.6% to 121.0%. This shows that the staining procedure did not noticeably influence the number of colonies visible at the end of the six-day incubation period. Figure 1 shows an example of microcolonies visible after fluorescent staining (left) and an almost identical pattern of colonies after re-incubation (right), demonstrating that the method is non-destructive. The three beer samples spiked with Dekkera bruxenlensis were incubated on Wallerstein agar at 27°C for two and for three days before staining. Following fluorescent micro-colony counting, the membranes were re-incubated. After a total incubation period of six days, the colonies visible to the eye were counted. To determine viability recovery, prepared plates were also incubated continuously for six days, whereupon the visible colonies were counted. The ‘Fluorescence count method’ columns in Table 2 reveal the fluorescence recovery rates determined for the three beer samples after two and three days of incubation. After two days, the fluorescence recoveries for Products A and C had reached the 70% threshold, but their fluorescence signals were too weak for reliable detection (see Figure 2). In addition, the fluorescence recovery for Product B was well below 70%, indicating that the colonies had not grown to the size required for fluorescence detection. After the three day incubation period, the fluorescence signals of all three beer samples were strong enough, and the fluorescence recovery rates all exceeded 70% (see Figure 3). All re-incubation recovery rates were also above 70%. This demonstrates that the detected micro-colonies went on to grow well after the staining procedure, becoming available for identification after a total of six days.
Conclusions
The results demonstrate that the fluorescent staining based micro-colony count method vastly reduces the time needed to quantitatively detect viable contaminants in soft drinks and beer. Compared with the traditional colony count method, detection of added yeasts, moulds and bacteria was completed 28 to 32 hours earlier – and thus up to three times faster – in the ten soft drink samples and 72 hours earlier in the three beer samples spoiled by a slow-growing yeast strain. The technique was shown to be compatible with the standard culture media traditionally used for the detection of contaminants in beverages. Neither the employed media nor any of the samples exhibited background fluorescence. Only two highly viscous fruit juice samples required pre-treatment to improve filterability. Because the method is non destructive, the fluorescent micro-colonies can be re-incubated to yield visible colonies, allowing identification of the contaminants using any available method. It can thus fully replace traditional colony count methods, enabling manufacturers’ quality control to reduce the time to result and thus to bring forward product release, reducing storage time and costs as well as the likelihood of spoiled products reaching the supermarket shelf. The technique can also help in the root cause analysis of process failures, providing additional data to increase the confidence in manufacturers’ process and quality control.
References
[1]Union of European Soft Drinks Associations, http://www.unesda.org/key-facts [2] Moore, J.E., McCalmont, M., Xu, J., Millar, B.C. and Heaney, N. "Asaia sp., an unusual spoilage organism of fruit-flavored bottled water." Applied and Environmental Microbiology 68 (2002), pp. 4130–4131. [3] Stratford, M. Food and Beverage Spoilage Yeasts, In: Querol, H. and Fleet, G. (eds.) Yeasts in Food and Beverages, Berlin, Germany: Springer-Verlag, Chapter 11 (2006), pp. 335–379. [4] Suzuki, K., Asano, S., Iijima, K., Ogata, T. and Kitagawa, Y. "Effects of Beer Adaptation on Culturability of Beer-Spoilage Dekkera/Brettanomyces Yeasts." J. Am. Soc. Brew. Chem. 66:4 (2008), pp. 239-244.
Contacts: Millipore SAS 39, route industrielle de la Hardt 67120 Molsheim France
Natalie Valton-El Khoury, Application Scientist Annabelle Caron PhD, Application Scientist Marilyn Vicente, Application Scientist, Head of the BioMonitoring Applications Group Renaud Chollet PhD Head of Biotechnologies and Reagents Tel: +33 3 90 46 87 33 E-mail: renaud.chollet@merckgroup.com





